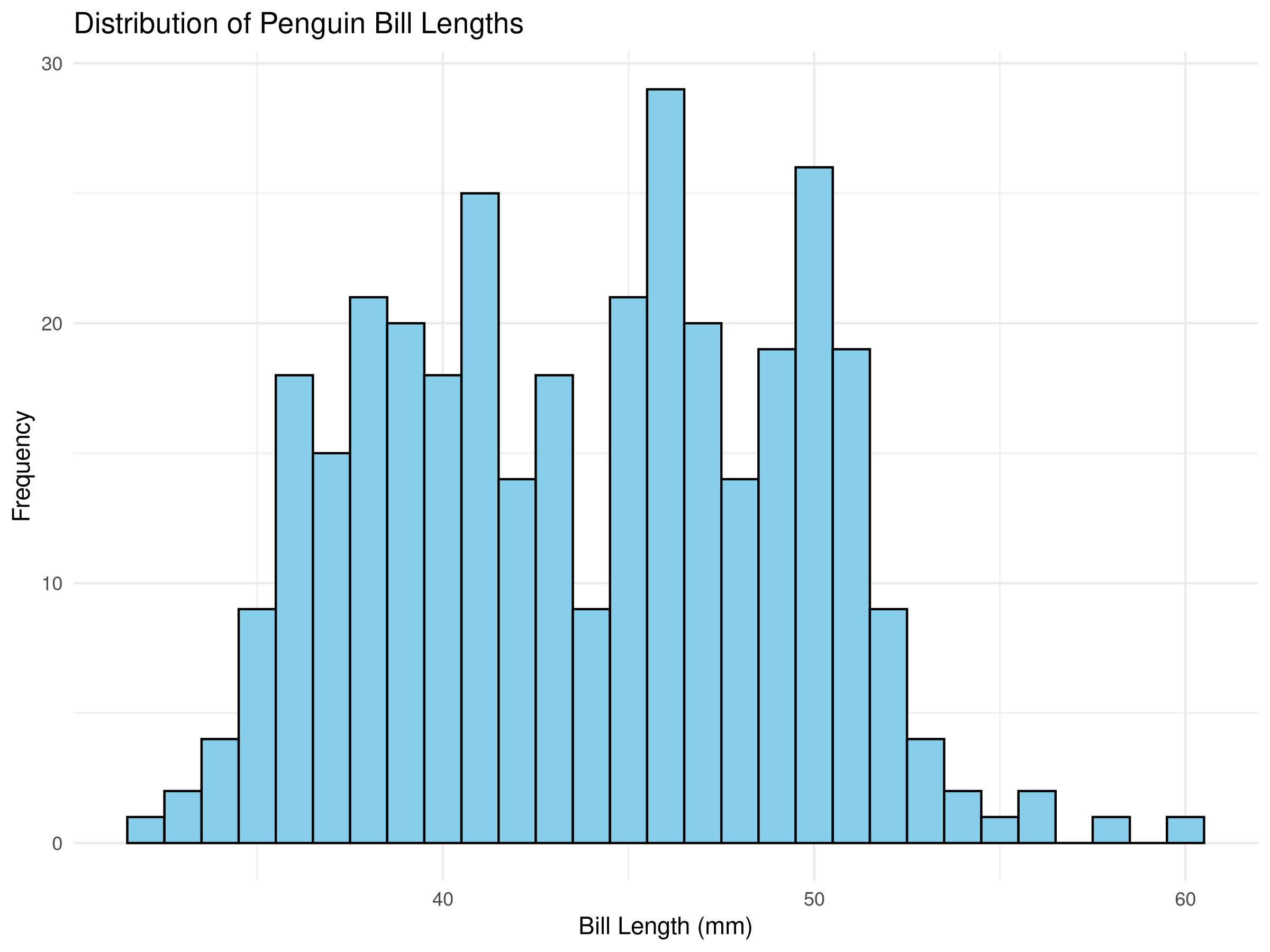
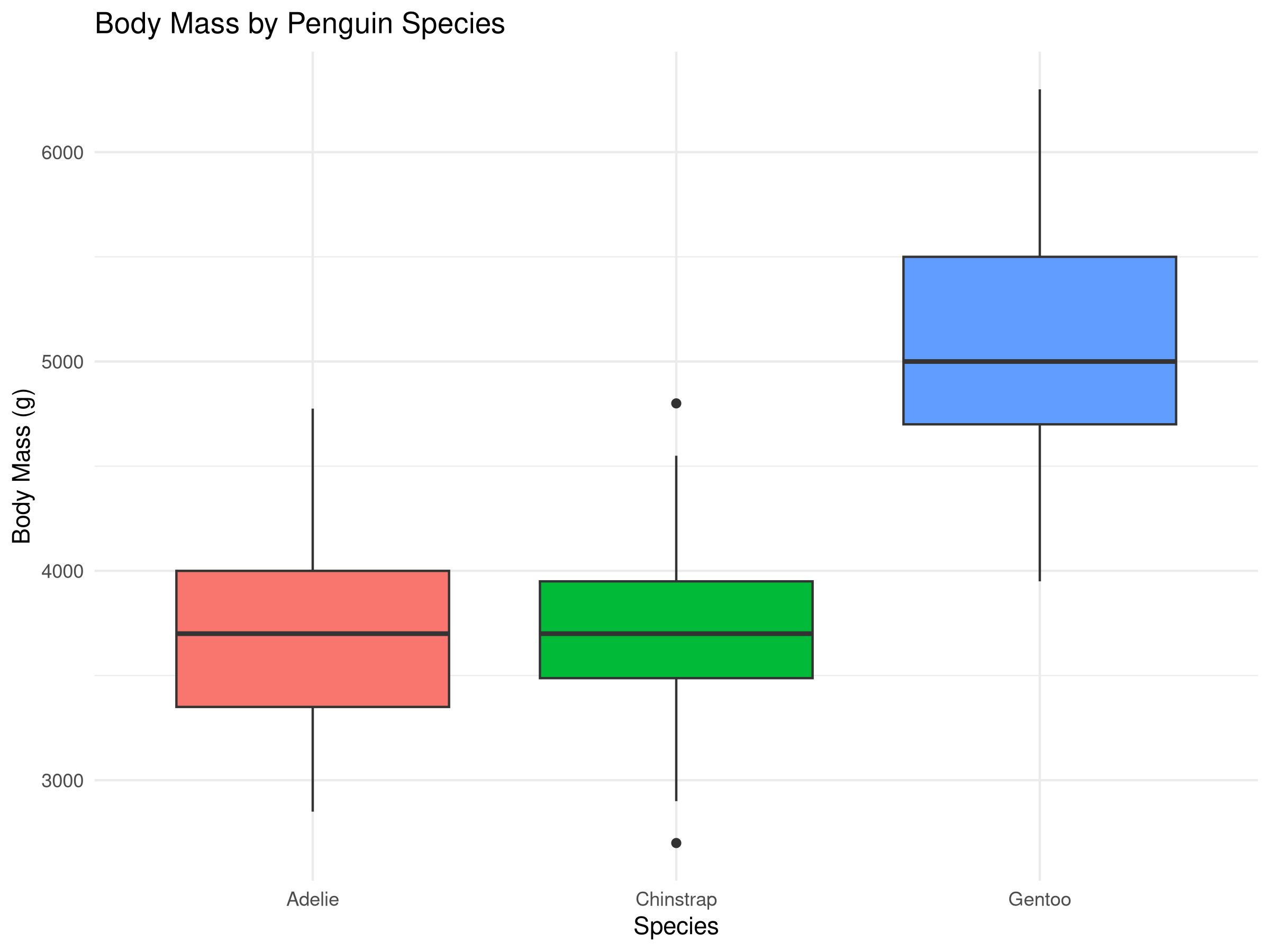
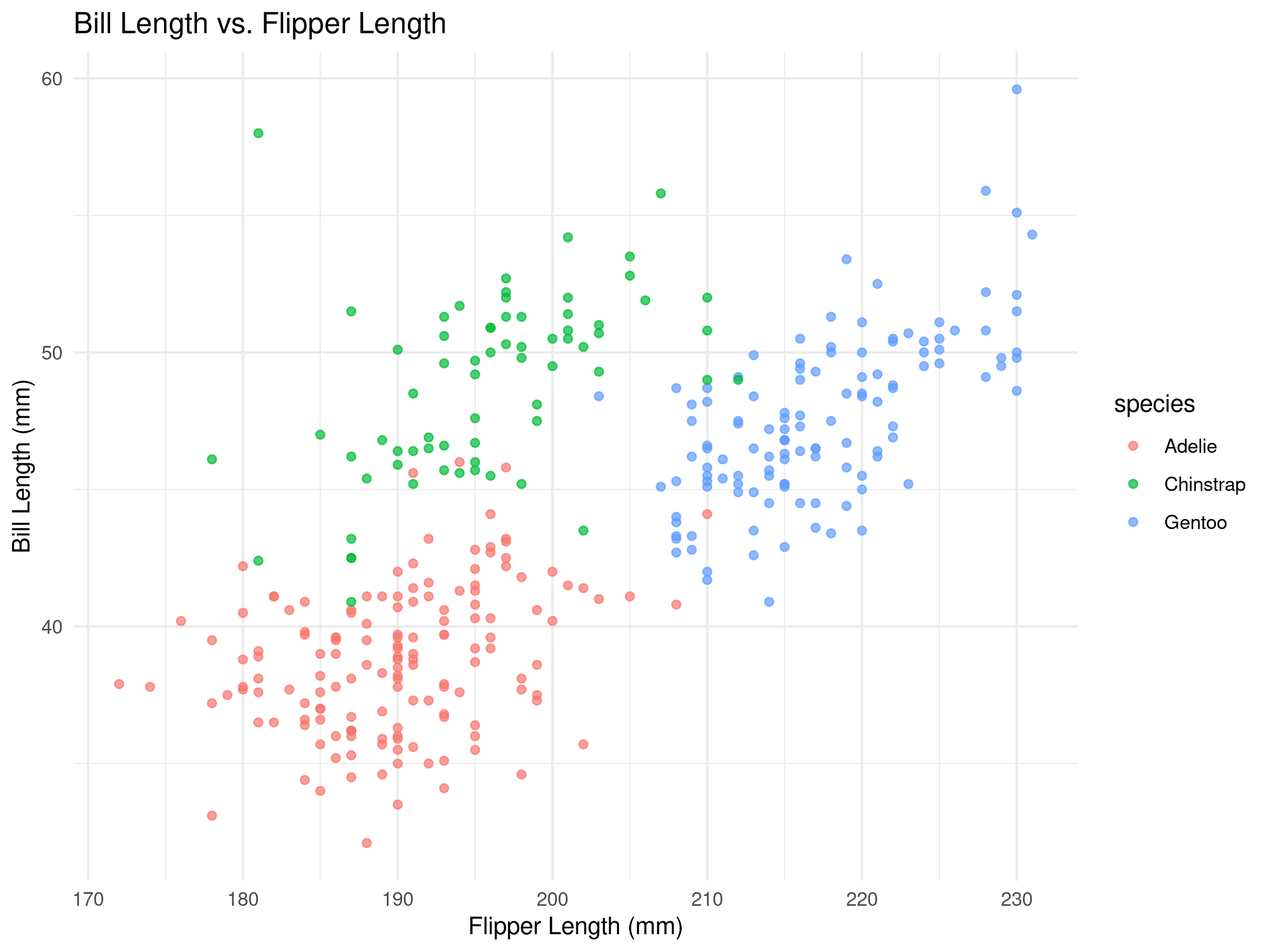
Rows: 500
Columns: 24
$ binomial_name <chr> "Abutilon pitcairnense", "Acaena exigua", "Acalypha …
$ country <chr> "Pitcairn", "United States", "Congo", "Saint Helena,…
$ continent <chr> "Oceania", "North America", "Africa", "Africa", "Oce…
$ group <chr> "Flowering Plant", "Flowering Plant", "Flowering Pla…
$ year_last_seen <chr> "2000-2020", "1980-1999", "1940-1959", "Before 1900"…
$ threat_AA <dbl> 0, 0, 0, 1, 1, 0, 0, 0, 0, 1, 0, 0, 1, 1, 0, 0, 0, 1…
$ threat_BRU <dbl> 0, 0, 0, 1, 0, 0, 0, 0, 0, 1, 1, 1, 1, 0, 1, 0, 0, 0…
$ threat_RCD <dbl> 0, 0, 0, 0, 1, 1, 1, 0, 0, 1, 0, 0, 0, 1, 0, 0, 0, 0…
$ threat_ISGD <dbl> 1, 1, 0, 1, 1, 0, 1, 1, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0…
$ threat_EPM <dbl> 0, 0, 1, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0…
$ threat_CC <dbl> 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0…
$ threat_HID <dbl> 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0…
$ threat_P <dbl> 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0…
$ threat_TS <dbl> 0, 0, 0, 0, 1, 0, 0, 0, 0, 1, 0, 0, 0, 0, 0, 0, 0, 0…
$ threat_NSM <dbl> 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0…
$ threat_GE <dbl> 1, 0, 0, 0, 0, 0, 0, 1, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0…
$ threat_NA <dbl> 0, 0, 0, 1, 0, 0, 0, 0, 1, 0, 0, 0, 0, 0, 0, 1, 1, 0…
$ action_LWP <dbl> 1, 0, 0, 0, 0, 0, 0, 1, 0, 0, 1, 0, 0, 0, 0, 0, 0, 0…
$ action_SM <dbl> 1, 0, 0, 0, 0, 1, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0…
$ action_LP <dbl> 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0…
$ action_RM <dbl> 1, 1, 0, 0, 0, 0, 0, 1, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0…
$ action_EA <dbl> 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0…
$ action_NA <dbl> 0, 0, 1, 1, 1, 0, 1, 0, 1, 1, 0, 1, 1, 1, 1, 1, 1, 1…
$ red_list_category <chr> "Extinct in the Wild", "Extinct", "Extinct", "Extinc…